This project is now in update mode. Check back regularly to see how things are progressing.
Tackling the Bubonic Plague
The Virtual Cures (a.k.a. Virtual Drug Screening) stream of the Freshman Research Initiative (FRI) at UT Austin seeks to discover new drugs against infectious diseases. The FRI engages undergraduates in authentic research experiences, which lead to increased student success in science while at UT. In our lab, they do this through drug discovery projects using both computational and wet-lab techniques as they build skills and contribute to the scientific body of knowledge. Led by Dr. Josh T. Beckham as the Research Educator in collaboration with Dr. Walt Fast of Pharmacy, students develop critical-thinking skills as they troubleshoot experiments and analyze data with the support of more senior, experienced peers (‘mentors’).

INFECTIOUS DISEASE
With the prevalence of disease burden across the world, especially in 3rd world countries, and the rise of anti-bacterial resistance, the need is evermore present to develop drugs to tackle these deadly bacteria and parasites in new ways. Past and current projects in the lab include investigating these pathogens/diseases: tuberculosis, malaria, Chagas disease (endemic to Central America), Listeriosis (Blue Bell Ice cream outbreak), and anthrax.
The students in the stream are now beginning new work on a particularly pressing infectious agent: Yersinia pestis – the bacterium that causes the bubonic plague which devastated the world in the middle ages (called the Black Death due to the patches of darkened skin from subsurface bleeding and the blackened fingers, noses, and toes of patients as the cells began to die). Unfortunately, this disease is re-emerging in some third world countries, has arisen in China this past November of 2019, and has popped up in Denver, Colorado - leading to evacuations of neighborhoods. While its re-emergence is threatening, it is even more daunting that the Yersinia pestis bacteria is a potential bioterrorist weapon. While there are a few current drugs, they are susceptible to resistance and do not work in all patients equally well. Finding new chemicals to bind to crucial proteins within these organisms can lead to new therapeutics to reduce the impact of these diseases on world health.
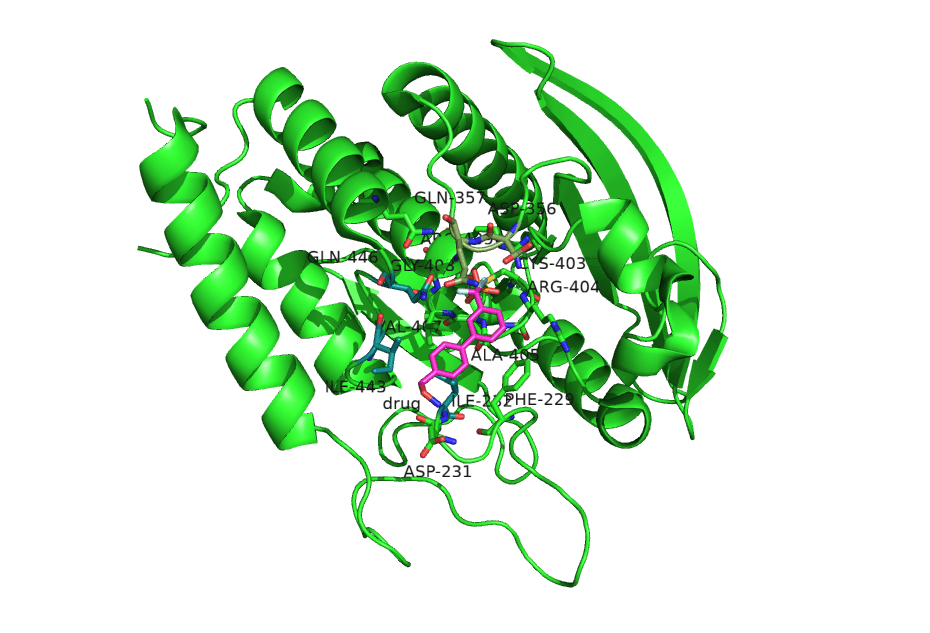
Figure 1. Structure of the YopH protein (PDB ID: 2Y2F) from Yersinia pestis - this protein helps the bubonic plague bacteria to evade the human's immune system
COMPUTATIONAL PREDICTIONS
In the Virtual Cures stream, we use computational techniques to help speed up this process and more accurately identify new chemical compounds that can be developed into drugs. If we can perform drug discovery faster and more effectively , the lives of many potential patients could be saved while also lowering the cost of the drugs. Often, we will screen 50,000 to 100,000 chemicals at a time on a high performance computer cluster with 72 computing threads to assess their ability to bind well to crucial proteins of the bacteria and parasites.
TESTING IN WET LAB
Once the most promising chemicals are identified from the computational work, the more labor-intensive work of validating them in the wet lab begins. To test if the chemicals actually bind, we need to do binding or enzyme assays. To do these assays, we need protein. But to make protein, we first need DNA. And so begins the long process of synthesizing our proteins for testing through the steps of recombinant DNA cloning, bacterial protein expression, purification, and characterization.

THE NEEDS OF THE LAB
These wet lab procedures require extensive equipment, reagents, and ‘human capital’. In particular, the Virtual Cures stream is requesting funds for:
- new chemicals so that we can test their ability to bind as drugs
- flourescent dye for binding assays
- necessary reagents for making protein (media components, Nickel affinity resins for purification)
- shaking incubator to grow up bacterial cells at 37 degrees Celsius.
- student support for a project leader
While several students in the lab will do the majority of work, we will fund a single student leader of the project and compensate them through a summer scholarship. Essentially, this student will be recognized as a leader among their peers and have the technical and organizational expertise to guide the other students through the project’s milestones.
- By giving your monetary support to the stream today you will be making this project a reality and provide invaluable research experiences to these great students!
Support us via credit card following the links to the right
Or Checks can be made out to: "The University of Texas at Austin - Tackling the Bubonic Plague" and delivered to us or mailed to: Annual Giving c/o Skyla Lowery Littlefield Carriage House 2410 Whitis Avenue Austin, TX 78712


$60
Students in the stream 2020
For this spring of 2020, the stream will welcome new students into the Freshman course and the upper-division ARI course.
$72
Threads on the BRCF Edu-POD
The Biomedical Research Computing Facility (BRCF) Edu-POD is a multiple processor computer cluster on which we run our molecular docking simulations (virtual screening).
$306
Number of chemicals
There are 306 chemicals in the 'CB306' library of compounds found from a large-scale screen of over 49,797 compounds used to find an inhibitor against Ricin toxin by the Robertus Lab (the original PI of the stream)
$527
Students since 2008
Since the stream's start in 2008 by Dr. Robertus and Dr. Kavanagh, we have engaged over 527 students in authentic research experiences
$2,238
Atoms in YopH protein
There are this many atoms in the Yersinia pestis protein YopH as determined in the crystal structure from the Protein Data Bank